 |
|
|
|
|
|
| |
|
 | |  |
|
 | Dr. Ananda K. Sarkar
Staff Scientist V
Phone: 91-11-26735220
Fax: 91-11-26741658
Email: aksarkar@nipgr.ac.in |
|
 | |  |
 Research Area Research Area |
| Plant Stem Cells and Architecture: Genetic, Epigenetic and Small RNA Mediated Regulation. |
 Academic Background Academic Background |
 |
Staff Scientist (2009 onwards): NIPGR |
 |
Ramalingaswami Fellow (2009 - 2014): NIPGR |
 | Ramalingaswami Fellow (2009): ICGEB (International Centre for Genetic Engineering and Biotechnology), New Delhi. |
 | Post Doc. Fellow (2006-2009): Cold Spring Harbor Laboratory (USA). |
 | Ph. D. (2001-2006): Albert-Ludwigs-Universitaet Freiburg (Germany). |
 | Res. Associate (2001): NRCPB (National Research Centre on Plant Biotechnology), IARI, New Delhi. |
 | M. Tech. (1998-2000): Indian Institute of Technology - Kharagpur. |
 | M. Sc. (1996-1998): Visva Bharati University (Shantiniketan). |
|
 Awards, Honors and Fellowships: (Click Here to Expand) Awards, Honors and Fellowships: (Click Here to Expand) |
 |
Ramalingaswami Re-Entry Fellowship (DBT, 2009- 2014). |
 | Prof. Archana Sharma Memorial Award (ISCA, 2010-2011). |
 | "Suma Cum Laude" (best grade award for Ph.D., University of Freiburg, Germany). |
 | Member of New York Academy of Science (2007-08) & AAAS, USA (2008-10). |
 | Postdoctoral fellowship (2006- 2009; at Cold Spring Harbor Lab, USA). |
 | Doctoral fellowship (2001- 2006) from German/European Research Foundations (DFG/SFB/REGIA). |
 | DBT fellowship (1998-1999) for M.Tech. (through GATE; at IIT- Kharagpur). |
 |
Academic Editor: Scientific Reports (Nature Publication Group) |
|
 Research Interests Research Interests |
| Genetic and epigenetic regulations of plant stem cells and architectures (root & shoot) |
| In higher plants, the continuous post-embryonic growth and lateral organ formation or branching is ultimately controlled by the root and shoot apical meristems through the activity of their respective "STEM CELL populations". Precise spatio-temporal regulation between diverse regulatory gene networks and their dynamic balance is quintessential for normal plant development. This balance is achieved by many layers of regulation at transcriptional, post-transcriptional and translational level. Epigenetic modifications play important role in gene transcription through DNA methylation and histone modifications. We are interested to understand the role of novel genes and epigenetic factors in regulation of plant stem cells, meristems, and patterning of root (and shoot) architecture. For this, we largely use molecular genetics, functional genomics and epigenetic work related techniques. |
| Role of small RNAs in plant stem cell regulation and root architecture |
| Besides protein coding genes, small non-coding RNAs (e.g. miRNA and siRNA) have recently been implicated in plant development and stress responses. We are exploring the role of miRNAs and other small RNAs in root meristem and architecture. We use NGS and microarray analysis to identify novel and relevant miRNAs, their targets. We further study their developmental role in plant stem cells or meristems, and root architecture through molecular genetic approaches. |
| Regulation of root architecture in crops (rice/maize) ‐ application and evolution |
The adaptability of higher plants to the changing environmental conditions as well as their productivity largely depends on their ability to modulate the patterning of root (and shoot). Therefore, we are exploring the functional conservation of regulation of root development between monocot crops and dicot model Arabidopsis. We are also studying the aforesaid regulations in rice/maize with the goal to improve root architecture and make them environmentally/economically more sustainable. We use transgenic and CRISPR/Cas9/Cpf1 based genome editing system to engineer and improve root architecture and stress/nutrient response.
Major techniques used: Students can learn/apply - basic DNA/RNA/Protein related works (including their isolation/ purification, gene cloning Nothern/ Western analysis. qRT-PCR etc.); ChIP, Y2H/protein interaction studies; advanced microscopy (light/ fluorescence, Confocal LSM, SEM, Laser Capture Microdissection etc.); microarray/ NGS for transcriptome/ miRNAome study; in situ localization of RNAs/ proteins; histology/ microtomy; molecular genetics, development of transgenic plants, mutational analysis, phenotyping etc. |
 Positions Available Positions Available |
| Roots and shoots are growing here; we welcome strong and efficient signaling candidates to contribute. Interested candidates, including NPDF, DBT-RA, CSIR-RA applicant with strong candidature may contact. |
 Group Members (Click Here to Expand) Group Members (Click Here to Expand) |
 |
Dr. Saikat Paul
Research Associate, CSIR
PhD from Calcutta University |
 |
Dr Ashutosh Kumar
CSIR-SRA (Scientists' Pool)
PhD from BIT, Mesra, Ranchi, Jharkhand |
 |
Dr. Sandeep Yadav
Research Associate (Ph. D. from Sarkar Lab NIPGR)
M.Sc. CCS HAU, Hissar, Haryana |
 |
Dr. Pramod Kumar
Research Associate (Ph. D. from Sarkar Lab NIPGR)
M.Sc. from CCS University, Meerut |
 |
Mr. Vishnu Mishra
Ph. D. Student
M.Sc. from BHU |
 |
Ms. Mahima
Ph. D. Student
M.S. from IISER - Mohali |
 |
Mr. Sourav Chatterjee
Ph. D. Student
M.S. from Calcutta University |
 |
Ms. Nidhi Gandhi
Ph. D. Student
M. Sc. from University of Rajasthan, Jaipur |
|
 Former Group Members (Click Here to Expand) Former Group Members (Click Here to Expand) |
 |
Dr. Shalini Mukherjee
National Postdoctoral Fellow (DST) Completed-2019
Ph.D. from University of Manitoba, Canada |
 |
Dr. Swati Verma
National Postdoctoral Fellow (DST) Completed-2019
PhD from IIT-Rurkee |
 |
Dr. Alka Singh
Research Associate (Sarkar Lab NIPGR)
Ph. D., CIMAP/ JNU |
 |
Dr. Vibhav Gautam
Research Associate, Ph. D. (Sarkar Lab NIPGR)
M.Sc. from University of Pune
Current Position: Faculty in BHU
|
 |
Dr. Archita Singh
Research Associate, Ph. D. (Sarkar Lab NIPGR)
M.Sc. from BHU, Varanasi
Current Position: NPDF at NRCPB |
 |
Dr. Sharmila Singh
Research Associate, Ph. D. (Sarkar Lab NIPGR)
M.Sc. from MSR University, Vadodara, PhD NIPGR
Current Position: NPDF at CCMB |
 |
Dr. Sharadha Roy
Research Associate, (Sarkar Lab NIPGR)
Ph. D. from Japan
Current Position: Publications |
 |
Mr. B. Rengasamy
Junior research fellow (JRF), (Sarkar Lab NIPGR)
|
 |
Ms. Oshin Sharma
Junior Research Fellow (JRF)
M.Sc. from Pune University |
|
 Selected Publications Selected Publications |
| |
Journal |
(5 Year)
Impact Factor |
Citation till September 2020 |
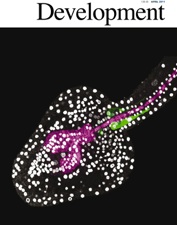
 Gautam V, Singh A, Yadav S, Singh S, Kumar P, Sarkar Das S, Sarkar AK*. Conserved LBL1-ta-siRNA and miR165/166-RLD1/2 modules regulate root development in maize. Development 2020. doi:10.1242/dev.190033. . Gautam V, Singh A, Yadav S, Singh S, Kumar P, Sarkar Das S, Sarkar AK*. Conserved LBL1-ta-siRNA and miR165/166-RLD1/2 modules regulate root development in maize. Development 2020. doi:10.1242/dev.190033. . |
DEVELOPMENT |
6.2 |
|
 Singh S, Singh A, Singh A, Mahima, Yadav S, Bajaj I, Kumar S, Jain A, and Sarkar AK*. Role of chromatin modifiers in stem cell regulation and meristem maintenance in Arabidopsis. J Exp Bot. 2019 Dec 3. pii: erz459. doi: 10.1093/jxb/erz459. [Epub ahead of print].. Singh S, Singh A, Singh A, Mahima, Yadav S, Bajaj I, Kumar S, Jain A, and Sarkar AK*. Role of chromatin modifiers in stem cell regulation and meristem maintenance in Arabidopsis. J Exp Bot. 2019 Dec 3. pii: erz459. doi: 10.1093/jxb/erz459. [Epub ahead of print]..
|
 |
7.011 |
1 |
 Singh S, Yadav S, Singh A, Mahima, Singh A, Gautam V, Sarkar AK*. Auxin signaling modulates LATERAL ROOT PRIMORDIUM1 (LRP1) expression during lateral root development in Arabidopsis. Plant J. 2019 Sep 4. doi: 10.1111/tpj.14520. [Epub ahead of print]. Singh S, Yadav S, Singh A, Mahima, Singh A, Gautam V, Sarkar AK*. Auxin signaling modulates LATERAL ROOT PRIMORDIUM1 (LRP1) expression during lateral root development in Arabidopsis. Plant J. 2019 Sep 4. doi: 10.1111/tpj.14520. [Epub ahead of print].
*LRP1 interact with SHI/STY family members, and its overexpression negatively affect lateral root development. Our results suggest that auxin and histone deacetylation affect LRP1 expression, and it acts downstream of LR forming auxin response modules to negatively regulate LRP development by modulating auxin homeostasis in Arabidopsis.
|
 |
6.141 |
4 |
 Farhat S, Jain N, Singh N, Sreevathsa R, Dash P, Rai R, Yadav S, Kumar P, Sarkar A, Jain A, Singh NK, Rai V. CRISPR-Cas9 directed genome engineering for enhancing salt stress tolerance in rice. Semin Cell Dev Biol. 2019 May 7. pii: S1084-9521(18)30119-8. doi: 10.1016/j.semcdb.2019.05.003. [Epub ahead of print]. Farhat S, Jain N, Singh N, Sreevathsa R, Dash P, Rai R, Yadav S, Kumar P, Sarkar A, Jain A, Singh NK, Rai V. CRISPR-Cas9 directed genome engineering for enhancing salt stress tolerance in rice. Semin Cell Dev Biol. 2019 May 7. pii: S1084-9521(18)30119-8. doi: 10.1016/j.semcdb.2019.05.003. [Epub ahead of print]. |
 |
6.629 |
5 |
 Kumar A, Singh A, Kumar P, Sarkar AK*. GAST proteins evolved through successive conjugation of novel motifs and their sub-functionalization. Plant Physiol. 2019 Jun; 180(2):998-1012. doi: 10.1104/pp.19.00305. Epub 2019 Apr 10. PMID: 30971449. Kumar A, Singh A, Kumar P, Sarkar AK*. GAST proteins evolved through successive conjugation of novel motifs and their sub-functionalization. Plant Physiol. 2019 Jun; 180(2):998-1012. doi: 10.1104/pp.19.00305. Epub 2019 Apr 10. PMID: 30971449.
*We have identified GASTs across plant kingdom and their four conserved novel motifs, which first originated in pteridophyte and then evolved through successive conjugation and sub-functionalization. AtGAST10 (Arabidopsis) and OsGASR9 (rice) homologues show structural and functional conservation in seed germination, and possibly act in hormone signaling.
|
 |
6.42 |
1 |
 Singh A, Gautam G, Singh S, Sarkar Das S, Verma S, Mishra V, Mukherjee S, and Sarkar AK*. Plant small RNAs: advancement in the understanding of biogenesis and role in plant development. Planta 2018 July 2, https://doi.org/10.1007/s00425-018-2927-5. Singh A, Gautam G, Singh S, Sarkar Das S, Verma S, Mishra V, Mukherjee S, and Sarkar AK*. Plant small RNAs: advancement in the understanding of biogenesis and role in plant development. Planta 2018 July 2, https://doi.org/10.1007/s00425-018-2927-5.
* Invited Review illustrating role of miRNAs & ta-siRNAs in the development of shoot, root, embryos, seed (incl. germination), and leaf in monocot and dicot plants. We expect it to be a good reference for small RNA researchers, teachers and students. |
 |
3.687 |
34 |
 Singh A, Kumar P, Gautam V, Rengasamy B, Adhikari A, Udayakumar M and Sarkar AK*. Root transcriptome of two contrasting indica rice cultivars uncovers regulators of root development and physiological responses. Sci. Rep. 2016 Dec 21;6:39266. doi: 10.1038/srep39266.. Singh A, Kumar P, Gautam V, Rengasamy B, Adhikari A, Udayakumar M and Sarkar AK*. Root transcriptome of two contrasting indica rice cultivars uncovers regulators of root development and physiological responses. Sci. Rep. 2016 Dec 21;6:39266. doi: 10.1038/srep39266..
*This paper describes molecular and physiological differences between two indica rice cultivars with stable and contrasting root system architecture (RSA), which we used as model system and identified many potential regulators of RSA and associated physiological responses (such as draught, salinity and nutrient availability) using functional genomics approach. |

Nature Publication |
4.576 |
13 |
 Rai V, Sanagala R, Sinilal B, Yadav S, Sarkar AK, Dantu PK, Jain A. Iron Availability Affects Phosphate Deficiency-Mediated Responses, and Evidences of Cross Talk with Auxin and Zinc in Arabidopsis. Plant Cell Physiol. 2015 Jun;56(6):1107-23. doi: 10.1093/pcp/pcv035. Rai V, Sanagala R, Sinilal B, Yadav S, Sarkar AK, Dantu PK, Jain A. Iron Availability Affects Phosphate Deficiency-Mediated Responses, and Evidences of Cross Talk with Auxin and Zinc in Arabidopsis. Plant Cell Physiol. 2015 Jun;56(6):1107-23. doi: 10.1093/pcp/pcv035.
*It uncovers the cross-talk between neutrients (Pi and Fe) and hormone (auxin) signalling in root development.
|
 |
4.799 |
40 |
 Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development. 2004 Feb;131(3):657-68. Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development. 2004 Feb;131(3):657-68.
*This article describes (for the FIRST time) novel WOX (WUSCHEL-RELATED HOMEOBOX) gene family, their in situ expression dynamics, and role in embryonic patterning events. WUS, the founder WOX, is the master regulator of SHOOT STEM CELLs. This work is also incorporated in "Plant Physiology" by Taiz & Zeiger, since 2010.
|
 |
6.192 |
704
(~47 /yr) |
 Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007 Apr 12;446(7137):811-4. Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007 Apr 12;446(7137):811-4.
* QC specific transcription factor WOX5 is required for maintenance of QC and root stem cells. It provides first molecular evidence for role of conserved regulators in shoot and root stem cell maintenance. This work is also incorporated in "Plant Physiology" by Taiz & Zeiger, since 2010.
|
 |
45.819 |
804
(~67 /yr) |
 Brooks L, Strable J, Elshire R, Zhang X, Ohtsu K, Sarkar AK, Hargreaves S., Eudy D., Pawlowska T, Nettleton D, Timmermans MCP, Schnable PS, Scanlon MJ. Microdissection of Shoot Meristem Functional Domains in Maize. PLoS Genet. 2009 May;5(5):e1000476. Brooks L, Strable J, Elshire R, Zhang X, Ohtsu K, Sarkar AK, Hargreaves S., Eudy D., Pawlowska T, Nettleton D, Timmermans MCP, Schnable PS, Scanlon MJ. Microdissection of Shoot Meristem Functional Domains in Maize. PLoS Genet. 2009 May;5(5):e1000476.
*Using Laser Capture Microdissection (LCM)-Microarray approach, it describes maize shoot meristem domain or organ specific gene expression signature and indicates function.
|
 |
5.03 |
72 |
 Douglas RD, Wiley D, Sarkar AK, Springer N, Timmermans MCP and Scanlon MJ. ragged seedling2 Encodes an ARGONAUTE7-Like Protein Required for Mediolateral Expansion, but Not Dorsiventrality, of Maize Leaves. Plant Cell. 2010 May;22(5):1441-51. Douglas RD, Wiley D, Sarkar AK, Springer N, Timmermans MCP and Scanlon MJ. ragged seedling2 Encodes an ARGONAUTE7-Like Protein Required for Mediolateral Expansion, but Not Dorsiventrality, of Maize Leaves. Plant Cell. 2010 May;22(5):1441-51.
*It demonstrates role of small RNA (tasi-RNA) in maize leaf development.
|
 |
9.618 |
69 |
 Singh A, Singh A, Panigrahi KC, Reski R and Sarkar AK. Balanced activity of microRNA166/165 and its target transcripts from the class III homeodomain leucine-zipper family regulates root growth in Arabidopsis thaliana. Plant Cell Rep. 2014 Jun;33(6):945-53. Singh A, Singh A, Panigrahi KC, Reski R and Sarkar AK. Balanced activity of microRNA166/165 and its target transcripts from the class III homeodomain leucine-zipper family regulates root growth in Arabidopsis thaliana. Plant Cell Rep. 2014 Jun;33(6):945-53.
|
 |
3.825 |
53 |
 Barik S, SarkarDas S, Singh A, Gautam V, Kumar P, Majee M, Sarkar AK. Phylogenetic analysis reveals conservation and diversification of miR166 genes among diverse plant species. Genomics. 2014 Jan;103(1):114-21. doi:10.1016/j.ygeno.2013.11.004. Barik S, SarkarDas S, Singh A, Gautam V, Kumar P, Majee M, Sarkar AK. Phylogenetic analysis reveals conservation and diversification of miR166 genes among diverse plant species. Genomics. 2014 Jan;103(1):114-21. doi:10.1016/j.ygeno.2013.11.004.
* This shows the evolution of miR166 and provides first evidence of Polycistronic miRNA in plant (Rice). |
 |
4.149 |
39 |
|  Other Publications Other Publications |
 |
Singh A, Gandhi N, Mishra V, Yadav S, Rai V, and Sarkar AK*. Role of abiotic stress responsive miRNAs in Arabidopsis root development. J. Plant Biochem. Biotechnol. 2020 Sep 10. https://doi.org/10.1007/s13562-020-00626-0. |
 |
Kumari S, Yadav S, Patra D, Singh S, Sarkar AK, Panigrahi KCS. Uncovering the molecular signature underlying the light intensity-dependent root development in Arabidopsis thaliana. BMC Genomics. 2019 Jul 20;20(1):596. doi: 10.1186/s12864-019-5933-5. |
 |
Verma S, Gautam V, and Sarkar AK*. Improved laser capture microdissection (LCM)-based method for isolation of RNA, including miRNA and expression analysis in woody apple bud meristem. Planta. 2019 Apr 11. doi: 10.1007/s00425-019-03127-0. [Epub ahead of print]. PMID: 30976910 |
 |
Gautam V, Singh A, Verma S, Singh S, Chatterjee S, Sarkar AK. (2019) Whole mount in situ localization of miRNAs and target mRNA transcripts in plants. 3 Biotech. 2019 May;9(5):193. doi: 10.1007/s13205-019-1704-x. Epub 2019 Apr 29. |
 |
Kumar A, Gautam V, Kumar P, Mukherjee S, Verma S and Sarkar AK. Identification and co-evolution pattern of stem cell regulator miR394s and their targets among diverse plant species. BMC Evo. Biol. 2019 Feb. 19:55; EVOB-D-18-00093.2. DOI: 10.1186/s12862-019-1382-7.
|
 |
Gautam V, Singh A, Verma S, Kumar A, Kumar P, Mahima M, Singh S, Mishra V, and Sarkar AK*. Role of miRNAs in root development of model plant Arabidopsis thaliana. Indian J Plant Physiol. 2017 Dec. 22 (4): 382-392. DOI:10.1007/s40502-017-0334-8. (RG IF: ~1.0). |
 |
SarkarDas S, Yadav S, Singh A, Gautam V, Sarkar AK, Nandi AK, Karmakar P, Majee M, and Sanan Mishra N. Expression dynamics of miRNAs and their targets in seed germination conditions reveals miRNA-ta-siRNA crosstalk as regulator of seed germination. Sci. Rep. 2017 Dec. DOI:10.1038/s42598-017-18823-8. IF (5 yrs): ~5.0. |
 |
Singh A, Roy S, Singh S, Sarkar Das S, Gautam V, Yadav S, Singh A, Samantha S, & Sarkar AK. Phytohormonal crosstalk modulates the expression of miR166/165s, target Class III HD-ZIPs, and KANADI genes during root growth in Arabidopsis thaliana. Sci. Rep. 2017 Jun 13;7(1):3408. doi: 10.1038/s41598-017-03632-w.
|
 |
Singh S, Singh A, Yadav S, Gautam V, Singh A, Sarkar AK*. Sirtinol, a Sir2 protein inhibitor, affects stem cell maintenance and root development in Arabidopsis thaliana by modulating auxin-cytokinin signaling components. Sci. Rep. 2017 Jan. doi: 10.1038/srep42450 (2017). |
 |
Gautam V, Singh A, Singh S, Sarkar AK. An Efficient LCM-Based Method for Tissue Specific Expression Analysis of Genes and miRNAs. Sci. Rep. 2016 Feb 10;6:21577. doi: 10.1038/srep21577. |
 |
Barik S, Kumar A, SarkarDas S, Yadav S, Gautam V, Singh A, Singh S and Sarkar AK. Coevolution Pattern and Functional Conservation or Divergence of miR167s and their Targets across Diverse Plant Species. Sci. Rep. 2015 Oct. 5:14611. DOI: 10.1038/srep14611. |
 |
Kumar A and Sarkar AK*. Apple CALCINEURIN B-LIKE PROTEIN10 genes have evolved to be novel targets of miR167s through sequence variation. Current Sci. 2017 Jan 10, 112 (1). doi: 10.18520/cs/v112/i01/147-150. |
 |
Gupta A, Sarkar AK, Senthil-Kumar M. Global Transcriptional Analysis Reveals Unique and Shared Responses in Arabidopsis thaliana Exposed to Combined Drought and Pathogen Stress. Front Plant Sci. 2016 May 24;7:686. doi: 10.3389/fpls.2016.00686. eCollection 2016. |
 |
Gautam V, Sarkar AK. Laser Assisted Microdissection, an Efficient Technique to Understand Tissue Specific Gene Expression Patterns and Functional Genomics in Plants. Mol Biotechnol. 2014 Nov 18. doi: 10.1007/s12033-014-9824-3. |
 |
Kaur C, Mustafiz A, Sarkar AK, Ariyadasa TU, Singla-Pareek SL, Sopory SK. Expression of abiotic stress inducible ETHE1-like protein from rice is higher in roots and is regulated by calcium. Physiol Plant. 2014 Sep;152(1):1-16. doi: 10.1111/ppl.12147. |
 | Singh S, Singh A, Roy S and Sarkar AK. SWP1 negatively regulates lateral root initiation and elongation in Arabidopsis. Plant Signal Behav. 2012 Dec;7(12):1522-5. doi: 10.4161/psb.22099. |
 | Deyhle F, Sarkar AK, Tucker EJ, Laux T. WUSCHEL regulates cell differentiation during anther development. Dev Biol. 2007 Feb 1;302(1):154-9. |
 | Nogueira FT, Sarkar AK, Chitwood DH, Timmermans MC. Organ Polarity in plants is specified through the opposing activity of two distinct small regulatory RNAs. Cold Spring Harb Symp Quant Biol. 2006;71:157-6. |
 | Pathre UV, Sarkar AK and Nath P. Modulation of Sucrose-Phosphate-Synthase Activity by Glucose-6-phosphate and Inorganic Phosphate during Post harvest Ripening in Banana. J. Plant Biol., 2001; Dec; vol. 28 (3), pp 301-305. |
|
 Book Chapter Book Chapter |
 |
Yadav S, Sarkar Das S, Kumar P, Mishra V and Sarkar AK. Tweaking microRNA-mediated gene regulation for crop improvement (chapter 3).Book: Advancement in Crop Improvement Techniques.2020, 26 June, Page 45-66. WoodHead Publishing, Elsevier Inc. UK. doi:10.1016/B978-0-12-818581-0.00003-6. ISBN: 978-0-12-818581-0. |
 |
Gautam V, Singh A, Singh S, Verma S, Sarkar AK*. (2019) Improved Method of RNA Isolation from Laser Capture Microdissection (LCM)-Derived Plant Tissues. In: Chekanova J.A., Wang HL.V. (eds) Plant Long Non-Coding RNAs. Methods in Molecular Biology, vol 1933. Humana Press, New York, NY. Methods Mol Biol. 2019;1933:89-98. doi: 10.1007/978-1-4939-9045-0_5. Print ISBN: 978-1-4939-9044-3; Online ISBN: 978-1-4939-9045-0. |
 |
Sarkar AK, Karthikeyan M, Gautam V, Barik S and SarkarDas S. Improving the Plant Root System Architecture to Combat Abiotic Stresses Incurred by Global Climate Changes. CLIMATE CHANGE AND ABIOTIC STRESS TOLERANCE. Wiley Wiley-VCH Verlag GmbH & Co. Weinheim, Germany, 2014. |
|
|
|
|
| |
|
|
|
|
|
|
 |
|